applying ml to study human genome¶
It never ceases to amaze me how our scientific communities around the globe continues to be so heavily vested in the study of human genome project! I've always been profoundly curious about this field. The inspiration to pursue this topic further stems from a couple of factors.
inspiration¶
- Quantitative Genetics paved my curiosity as a grad student.
- Reading "Code Breaker" from Isaac Walterson's inspirational and fantastic storytelling of Dr. Doudna - her incessant quest to develop ways to help humans treat disease by developing the famous CRISPR gene editing instrument. This was pretty influential for me to dive deeper in this subject and further understand how DNA sequencing, particularly mRNA which was the focus of Dr. Doudna's pioneering work that inspires me to learn more.
- Genetics has always been in the back of mind, and a passionate topic. So this project was a no-brainer.
In my creative attempt to dive into machine learning, this is it - right here!
It is absolutely not a new topic for me.
So, with this blog, I knew I had write something about human genetics and here it goes!
examining telomeric DNA sequences¶
Chromosomes are structures that live in our cell's nucleus. They're are made of protein and DNA molecules that carry our genomic information from cell to cell. We have 22 pairs of these numbered chromosomes (autosomes) and one pair of sex chromosomes (XX or XY), for a total of 46. Each pair contains two chromosomes, one coming from each parent, which means that children inherit half of their chromosomes from their mother and half from their father. Chromosomes can be seen through a microscope when the nucleus dissolves during cell division.
Here's a great video that explains this 3D!
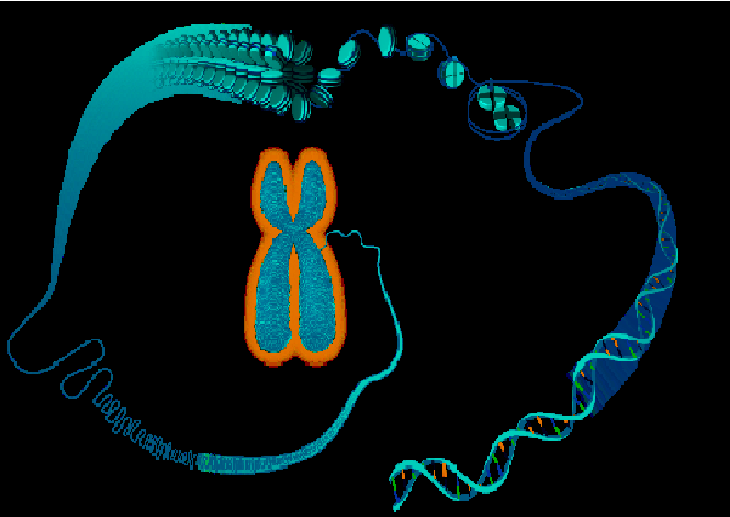
A telomere is a region of repetitive short DNA sequences at the end of a chromosome that gets repeated hundreds of times. Their complete replication and stability is essential for the human genome. Its job is to protect the ends of our chromosomes from becoming too frayed or tangled up! It's essentially a really long piece of DNA that is comprised of two loose ends as shown on the image here. These are called T-loops that help keep chromosome from clinging onto each other. And so every time a cell dividies, it gets cut off, until a telomeric enzyme called "telomerase" help adds those repeats back into the ends of the chromosomal strand to prevent it from becoming too short. Each time a cell divides, telomeres keeps shrinking until the cell can no longer divides, our cell dies.
Telomeres' progressive length shortening is a key biomarker for age-associated diseases in our chromosome. There have been substantial challenges even with quantifying evidences that resulted from lack of corrected method used to correctly measure its length in many research studies. With this being said, this project is to further dive into different datasets that have both supported and refuted this biomarker as age-associated link to aging by analyzing, predicting and building machine learning models to validate its accuracy and consistency of such claims.
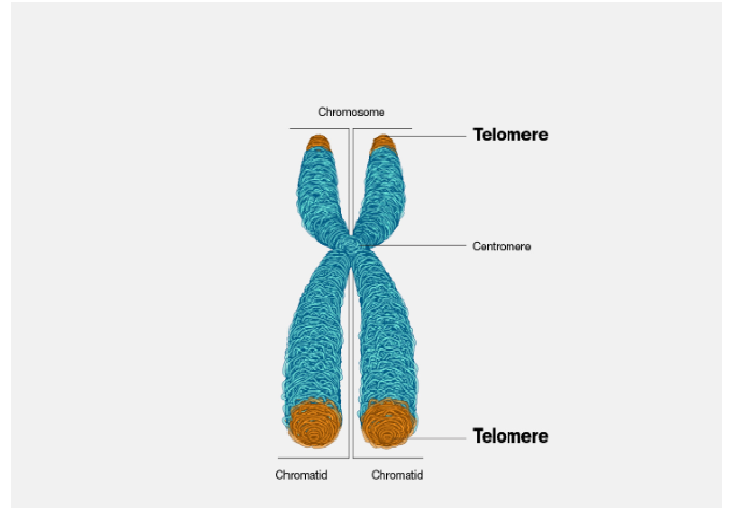
source: National Human Genomic Institute
multivariate factorial analysis¶
I wanted to do a meta analysis of combing through several research to study if human telomeres are affected by their upbringings via nature vs nurture that help us live longer. I collected several datasets that refuted and supported both claims that nature and nurture do in fact affect our telomeric DNA sequences.
Essentially, the scientific question to ask from the first dataset is: Deriving from the parents's chromosomal DNA sequencing structure - do the telomeres lengthen or shorten due to age and time as a repercussion of their upbringing or DNA genetic inheritance? Based on this study, the researchers concluded that from a longitudinal study of sparrow weavers that older parents do in fact propagated offsprings with longer telomeres, which yielded longer lifespan for their offsprings!
data exploration¶
sample analysis¶
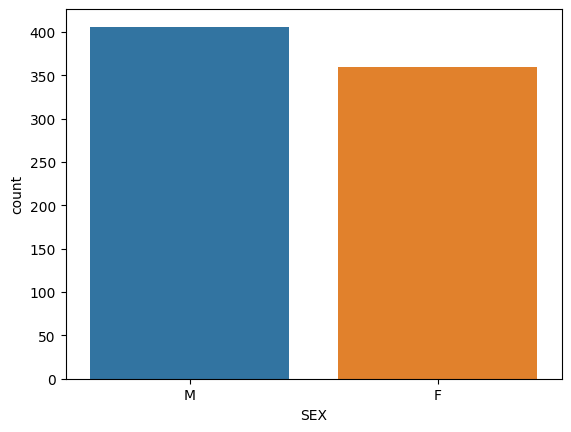
Sample population size for M and F for white-browed sparrow weavers.

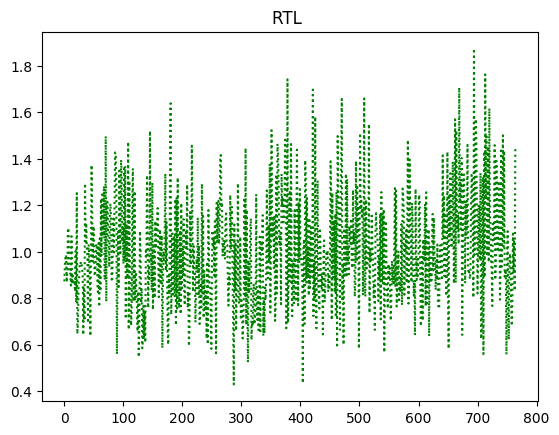
RTL as a function of time¶
As one can see that, duration and RTL is correlated here. This is examining the entire dataset without applying any ML models. This at least gives some exposure according to this dataset, that there is some evidence when observing both variables together.
supervised learning - linear regression¶
The simplest approach for me is to do a multivariate factorial analysis - generate a linear regression ml model to predict the effects of maternal vs paternal age as a function of time (days), if it's in fact affecting their offsprings' relative telomere length (RTL) in wild animals. This dataset is a collection of chromosomal datasets of wild animals containing telomeres traits, parental ages, dna extraction and sampling periods over a period of a few years; and comparing age as a function of time elapsed. The first dataset looks into wild white-browed sparrow weaver's breeding length.
Based on my analysis using a linear regression model, it is evident that maternal's influence in offsprings telomere length resulting in longer lifespan is due to age and duration. Particularly, when you're looking at the linear regression model I built applying both maternal and paternal's mean age as a function of time , the RTL's (relative telomere length measured by qPCR is a biomarker for physiological aging) has a higher R correlation in maternal vs paternal!
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
x = df["MATERNAL.AGE.DAYS"]
y = df["RTL"]
slope, intercept, r, p, std_err = stats.linregress(x, y)
def generate_linear_regression(x):
return slope * x + intercept
linregmodel = list(map(generate_linear_regression, x))
plt.scatter(x, y)
plt.plot(x, linregmodel, color = "red")
plt.gca().set_title("Linear regression model to predict RTL against maternal's age in days")
plt.show()
print(f"correlation = {r}, std error = {std_err}, slope = {slope}, intercept = {intercept}")
Looking at a simple linear regression model, it's clear that there is some significant correlation between parents' age and duration in DNA extraction and sampling period when it comes to comparing the maternal vs paternal DNA structural sequences and its influence in their offsprings life span, where their telomeric length for maternal is more signficant.
Maternal RTL based on age and duration
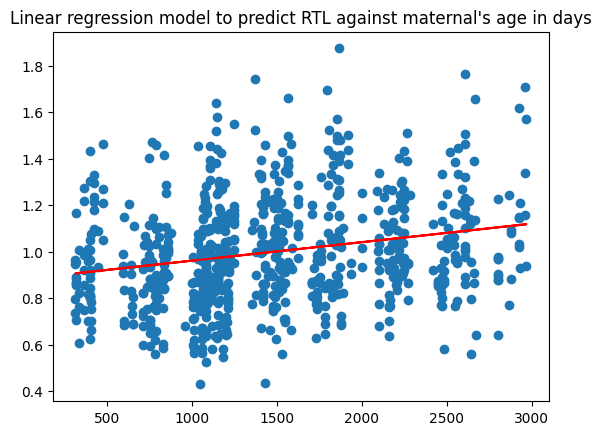
- correlation = 0.2302716811493559
- std error = 1.2200096429220074e-05
- slope = 7.974377994149696e-05
- intercept = 0.8813067639152621
Paternal's RTL based on age and duration
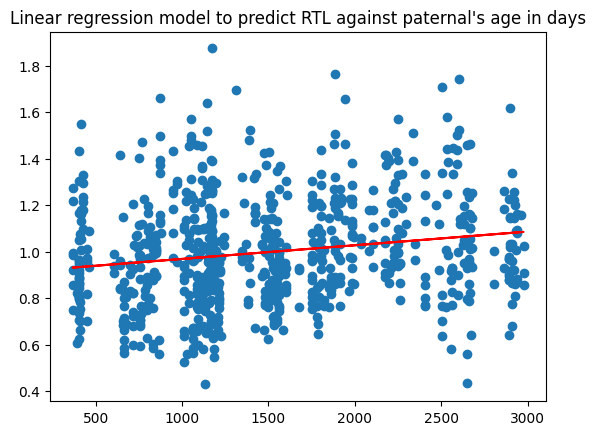
- correlation = 0.18167682726626644
- std error = 1.153510270254853e-05
- slope = 5.88669435519415e-05
- intercept = 0.9098460577841743